Note
Go to the end to download the full example code.
Trajectory Analysis¶
This example demonstrates how to analyze a polymer trajectory using pylimer-tools, including mean square displacement (MSD) and radius of gyration calculation.
import os
import matplotlib.pyplot as plt
import numpy as np
from pylimer_tools_cpp import UniverseSequence
# Load a LAMMPS dump trajectory file (replace with your file)
file_path = os.path.join(
os.getcwd(),
"../..",
"tests/pylimer_tools/fixtures/",
)
sequence = UniverseSequence()
sequence.initialize_from_dump_file(
initial_data_file=os.path.join(file_path, "big_dump_file_data.out"),
dump_file=os.path.join(file_path, "big_dump_file.lammpstrj"),
)
# Compute the mean square displacement for all atoms
msd = sequence.compute_msd_for_atoms(
atom_ids=[a.get_id() for a in sequence[0].get_atoms()],
nr_of_origins=50, # Number of origins to use for MSD calculation
)
You can also compute other properties like radius of gyration, end-to-end distance, etc. by using the “lazy” iterator over the sequence This allows you to process large trajectories without loading everything into memory at once.
# Compute radius of gyration over time
rgs = []
timesteps = []
for universe in sequence:
rgs.append(
np.mean(
[
m.compute_radius_of_gyration()
for m in universe.get_molecules(
-1
) # -1 because we don't have any crosslinks to filter out
]
)
)
timesteps.append(universe.get_timestep())
# Create a two-panel plot: MSD and Rg
fig, axs = plt.subplots(2, 1, figsize=(10, 10), sharex=False)
# Panel 1: MSD
axs[0].plot(
list(
msd.keys()), list(
msd.values()), label="Mean Square Displacement")
axs[0].axhline(
np.mean(
list(
msd.values())),
color="black",
linestyle="--",
label="Mean")
# add cumulative mean
axs[0].plot(
list(msd.keys()),
[np.mean(list(msd.values())[: i + 1]) for i in range(len(msd))],
label="Cumulative Mean",
color="red",
)
axs[0].set_xlabel("$\\tau$")
axs[0].set_ylabel("MSD")
axs[0].set_title("Mean Square Displacement")
axs[0].set(xscale="log", yscale="log")
axs[0].legend()
# Panel 2: Radius of Gyration
axs[1].plot(timesteps, rgs, label="Radius of Gyration", color="orange")
axs[1].axhline(np.mean(rgs), color="black", linestyle="--", label="Mean")
# add cumulative mean
axs[1].plot(
timesteps,
[np.mean(rgs[: i + 1]) for i in range(len(rgs))],
label="Cumulative Mean",
color="red",
)
axs[1].set_xlabel("Timestep")
axs[1].set_ylabel("Radius of Gyration")
axs[1].set_title("Radius of Gyration Over Time")
axs[1].set(xscale="log", yscale="log")
axs[1].legend()
plt.tight_layout()
plt.show()
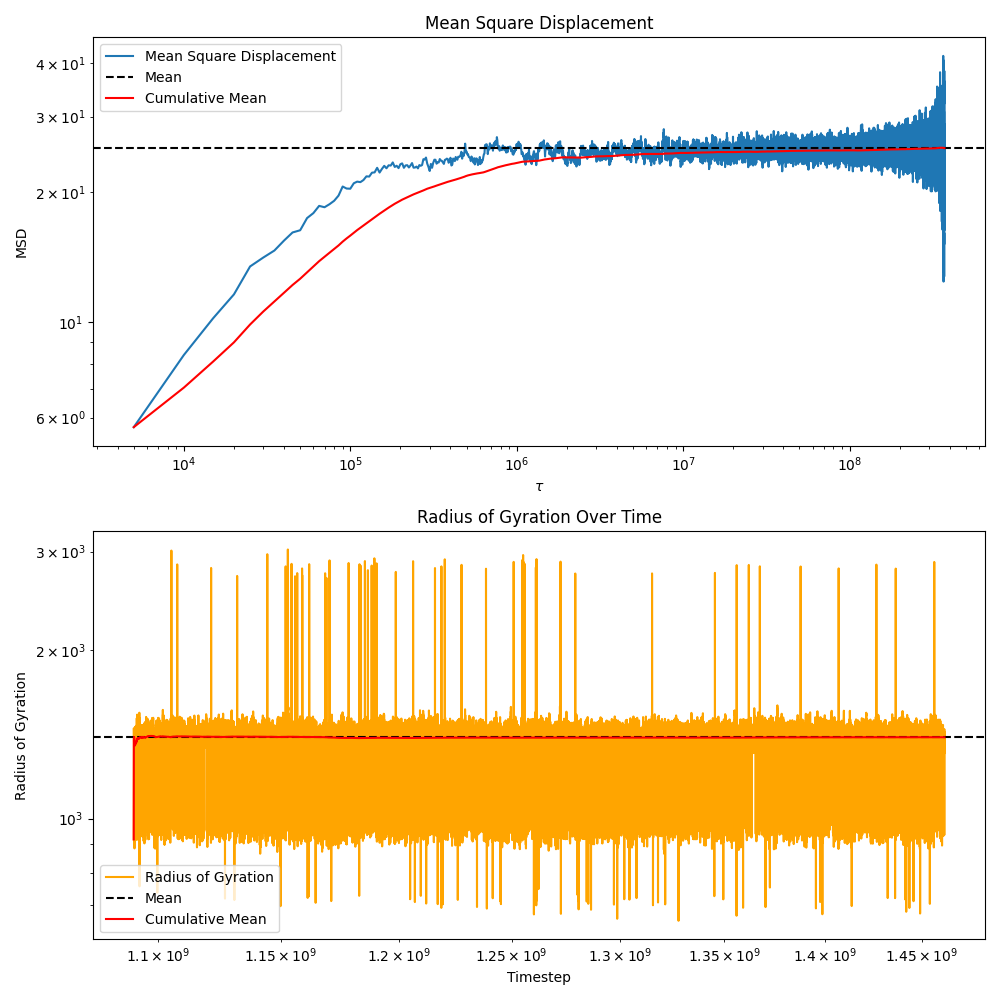
Please beware that the MSD is computed in an \(N^2\) manner, which can be slow for large systems, many origins and long trajectories.
If you want more origins or otherwise better performance,
consider using the tidynamics package instead like so:
import tidynamics
# using sorted atom ids to make sure the order is consistent
# across all timesteps, which is important for MSD calculation
sorted_atom_ids = sorted([a.get_id() for a in sequence[0].get_atoms()])
msd_result = tidynamics.msd(
np.array(
[
[
coord
for id in sorted_atom_ids
for coord in u.get_atom_by_vertex_id(id).get_coordinates()
]
for u in sequence
]
)
)
Total running time of the script: (9 minutes 47.636 seconds)