Note
Go to the end to download the full example code.
Monte Carlo Configuration¶
In the following, there are a few calls showing how to fine-tune the Monte Carlo network generation process:
import copy
import time
import matplotlib.pyplot as plt
from pylimer_tools.io.bead_spring_parameter_provider import (
ParameterType,
get_parameters_for_polymer,
)
from pylimer_tools_cpp import MCUniverseGenerator
generator = MCUniverseGenerator(
50.0, 50.0, 50.0
) # Create a generator for a 50x50x50 simulation box
# Configure the main options of the generator
generator.set_seed(12345) # Set random seed for reproducibility
generator.set_bead_distance(1.0) # Set mean bead-to-bead distance
# Configure whether to generate loops or not
generator.config_primary_loop_probability(0.0) # Primary loops prohibited
generator.config_secondary_loop_probability(0.0) # Secondary loops prohibited
# Improve performance of sampling pairs of crosslinks for large systems
# Does not sample all pairs of crosslinks anymore,
# only 99.9994% of those that are selected, but is ca. 60 times faster
generator.use_zscore_max_distance(3.29)
# Configure MC parameters
# If you do not trust the Brownian Bridge process we use,
# you can add additional Monte Carlo steps
generator.config_nr_of_mc_steps(5000) # More MC steps for better equilibration
In the following, we plot a performance comparison of using the z_score max distance or not, for a relatively small system. Please note that this always also depends on your system size, and even the density and length of the strands. If most distances have to be checked anyway, the overhead of the limit check would be apparent, for example.
Apart from the z-score, there are two more methods to configure the limit used:
- disable_max_distance()
- use_linear_max_distance()
# Get parameters for PDMS polymer density and bead distance
params = get_parameters_for_polymer(
"PDMS", parameter_type=ParameterType.GAUSSIAN)
n_strands = 50000
n_atoms_per_strand = 50
volume = (n_strands * n_atoms_per_strand + 0.5 *
n_strands) / params.get_bead_density()
generator_without_zscore = MCUniverseGenerator(
volume ** (1 / 3),
volume ** (1 / 3),
volume ** (1 / 3),
)
generator_without_zscore.set_seed(12345)
generator_without_zscore.set_bead_distance(1.0)
generator_without_zscore.add_crosslinkers(int(0.5 * n_strands), 4)
generator_without_zscore.add_strands(
n_strands, [n_atoms_per_strand for _ in range(n_strands)]
)
generator_without_zscore.disable_max_distance()
generator_with_zscore = copy.copy(generator_without_zscore)
start_time_without = time.time()
generator_without_zscore.link_strands_to_conversion(
crosslinker_conversion=0.925)
end_time_without = time.time()
print(
f"Time without max distance: {
end_time_without -
start_time_without: .2f} seconds"
)
generator_with_zscore.use_zscore_max_distance(3.0)
start_time_with = time.time()
generator_with_zscore.link_strands_to_conversion(crosslinker_conversion=0.925)
end_time_with = time.time()
print(
f"Time with z-score max distance: {end_time_with - start_time_with:.2f} seconds")
# plot the difference for the thumbnail
plt.figure()
plt.bar(
["Without Limit", "With Z-Score"],
[end_time_without - start_time_without, end_time_with - start_time_with],
color=["tab:blue", "tab:orange"],
)
plt.ylabel("Time (seconds)")
plt.show()
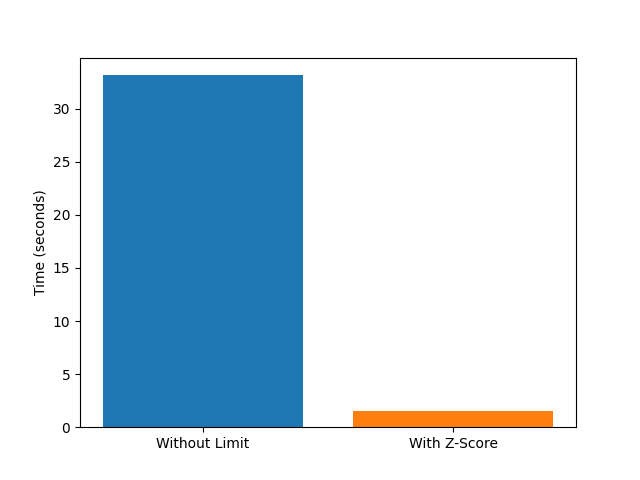
Time without max distance: 34.61 seconds
Time with z-score max distance: 11.27 seconds
Total running time of the script: (0 minutes 46.126 seconds)