Note
Go to the end to download the full example code.
Decompose/Split Structure¶
This example demonstrates how to decompose a crosslinked polymer network into chains and analyze its structure using pylimer-tools.
The following approaches are shown:
Removing crosslinkers and analyzing polymer strands
Decomposing the network to chains while keeping crosslinkers
Finding clusters after manual junction removal
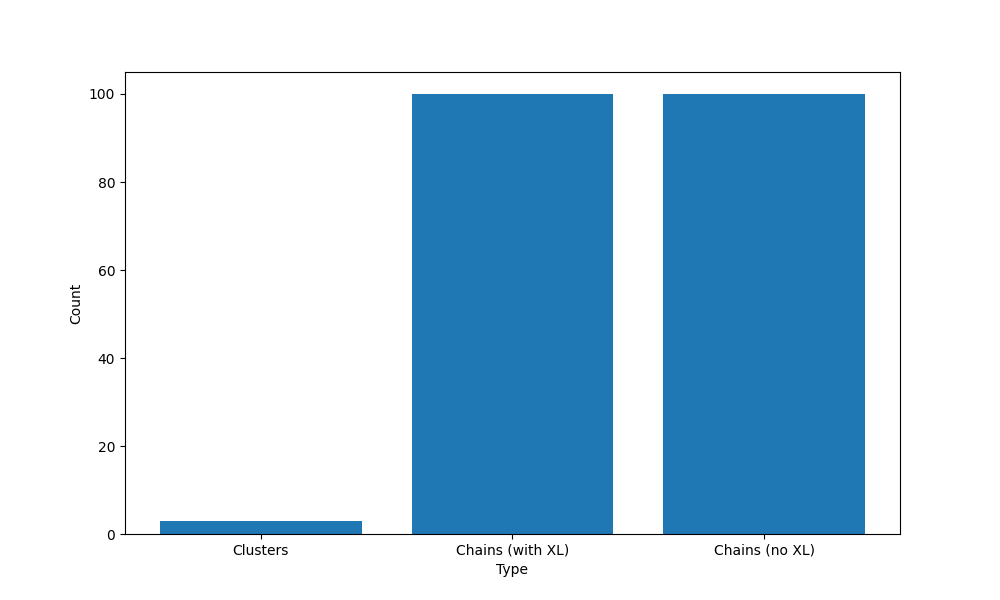
Number of chains (without crosslinkers): 100
Number of chains (with crosslinkers): 100
Number of clusters: 3
import os
import matplotlib.pyplot as plt
from pylimer_tools.io.read_lammps_output_file import read_data_file
from pylimer_tools_cpp import Universe
# Replace with your crosslinker type
crosslinker_type = 2
# Load your network (replace path accordingly)
universe = read_data_file(
os.path.join(
os.getcwd(),
"../..",
"tests/pylimer_tools/fixtures/structure/network_100_a_46.structure.out",
)
)
assert isinstance(universe, Universe)
# 1. Decompose by removing crosslinkers
chains = universe.get_molecules(crosslinker_type)
print(f"Number of chains (without crosslinkers): {len(chains)}")
# 2. Decompose keeping crosslinkers in all chains
chains_with_xl = universe.get_chains_with_crosslinker(crosslinker_type)
print(f"Number of chains (with crosslinkers): {len(chains_with_xl)}")
# 3. Find clusters (if you removed junctions yourself)
clusters = universe.get_clusters()
print(f"Number of clusters: {len(clusters)}")
# Plot the weights of the clusters, chains and molecules
plt.figure(figsize=(10, 6))
plt.bar(
["Clusters", "Chains (with XL)", "Chains (no XL)"],
[len(clusters), len(chains_with_xl), len(chains)],
)
plt.xlabel("Type")
plt.ylabel("Count")
plt.show()
Total running time of the script: (0 minutes 0.062 seconds)