Note
Go to the end to download the full example code.
Generate a Polydisperse Polymer Network¶
This example demonstrates how to use the MCUniverseGenerator
to create a polydisperse end-linked polymer network with varying strand lengths.
For strand length distribution, in this example, we show how to use the log-normal distribution as proposed by Tsimouri et al. [TCG21]. Alternatives were proposed as well, such as the inverse Gaussian or a ternary distribution.
import math
import random
import matplotlib.pyplot as plt
from pylimer_tools.io.bead_spring_parameter_provider import (
ParameterType,
get_parameters_for_polymer,
)
from pylimer_tools_cpp import MCUniverseGenerator
# Get parameters for PDMS polymer density, bead distance, and
# characteristic ratio
params = get_parameters_for_polymer(
"PDMS", parameter_type=ParameterType.GAUSSIAN)
C_inf = params.get_characteristic_ratio()
# setup strand lengths
n_strands = 1000
average_length = 50
polydispersity = 1.25
# Calculate parameters for log-normal distribution
s = math.sqrt(math.log(polydispersity))
m = math.log(average_length) - 0.5 * s**2
# sample strand lengths from log-normal distribution
strand_lengths = [int(random.lognormvariate(m, s)) for _ in range(n_strands)]
n_atoms_total = sum(strand_lengths) + 0.5 * n_strands # add crosslinker sites
volume = n_atoms_total / params.get_bead_density()
generator = MCUniverseGenerator(
volume ** (1 / 3),
volume ** (1 / 3),
volume ** (1 / 3),
)
generator.set_bead_distance(
params.get("<b>").to(params.get("distance_units")).magnitude
)
generator.add_strands(nr_of_strands=n_strands, strand_lengths=strand_lengths)
# 4-functional crosslinkers
generator.add_crosslinkers(
int(0.5 * n_strands), crosslinker_functionality=4, crosslinker_type=2
)
generator.link_strands_to_conversion(
crosslinker_conversion=0.925, # 92.5% of crosslinker sites used
)
universe = generator.get_universe()
universe.set_masses({1: 1.0, 2: 1.0}) # Set masses for LAMMPS
assert math.isclose(
universe.compute_polydispersity_index(2),
polydispersity,
rel_tol=0.1,
)
Refer to File I/O: Readers & Writers for how you can save the universe to a file.
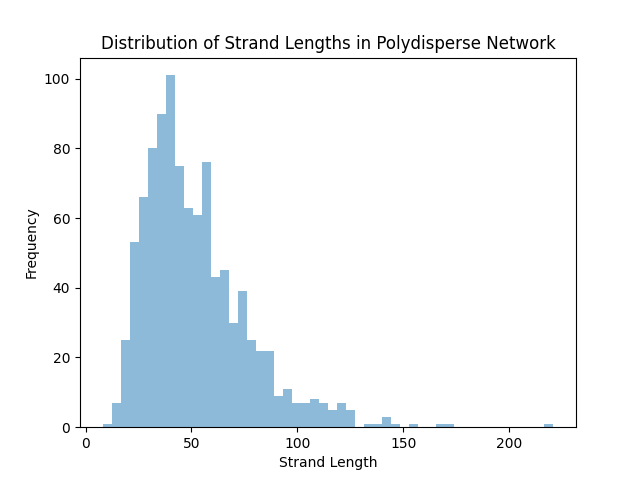
For now, we will show that this produces a variety of different strand lengths:
strand_lengths = [m.get_nr_of_atoms()
for m in universe.get_chains_with_crosslinker(2)]
# Plot the distribution of strand lengths
plt.figure()
plt.hist(strand_lengths, bins=50, alpha=0.5, label="Strand Lengths")
plt.xlabel("Strand Length")
plt.ylabel("Frequency")
plt.title("Distribution of Strand Lengths in Polydisperse Network")
plt.show()
Total running time of the script: (0 minutes 6.576 seconds)