Structure Analysis¶
pylimer-tools offers various tools to manipulate and analyze the topology of polymer networks.
Here you can find examples of some of the functionality provided by the package.
Decomposing Crosslinked Networks to Chains¶
There are a number of ways how a given Universe can be decomposed.
The most common use case is to decompose a crosslinked network into its constituent chains, which can then be used for further analysis or processing.
To find the chains, there are two main approaches:
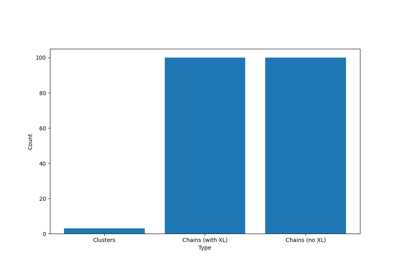
- Removing the crosslinkers and analysing the remaining polymer strands using get_molecules()
- Decomposing the network to chains, but keeping the crosslinkers in place, possibly being repeated in all the chains it is associated with. This decomposition can be done using the get_chains_with_crosslinker() method.
pylimer-tools provides two more advanced methods for decomposing networks:
- If you removed the junctions yourself, you can use get_clusters()
If you want even more control, you could assemble your own decomposition algorithm using the Universe class and its methods, such as get_atoms_connected_to().
Finding Loops¶
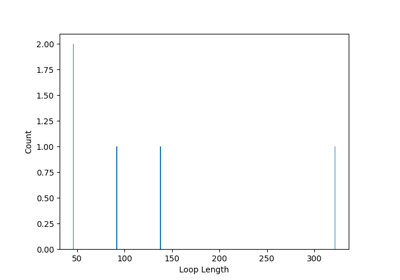
You can also find loops in the network using the find_loops() method.
If you want to count the number of loops present in the network, as a function of their length, you can use the count_loop_lengths() method.
Finally, to find the shortest loop a specific atom is involved in, you can use the find_minimal_order_loop_from() method.
Caution
There are exponentially many paths between two crosslinkers of a network, and you may run out of memory when using these functions, if your network is lattice-like.