Note
Go to the end to download the full example code.
Generate Vulcanized Networks¶
In this example, we create a vulcanized network using the pylimer-tools library.
import os
import matplotlib.pyplot as plt
import numpy as np
from scipy.stats import chisquare, kstest
from pylimer_tools.io.bead_spring_parameter_provider import (
ParameterType,
get_parameters_for_polymer,
)
from pylimer_tools_cpp import (
DataFileWriter,
MCUniverseGenerator,
randomly_sample_entanglements,
)
# Get parameters for PDMS polymer density and bead distance
params = get_parameters_for_polymer(
"PDMS", parameter_type=ParameterType.GAUSSIAN)
n_strands = 10000
n_atoms_per_strand = 50
# Determine the required volume for this many strands
# and this type of polymer
volume = n_strands * n_atoms_per_strand / params.get_bead_density()
# Create generator for simulation box corresponding to the volume
generator = MCUniverseGenerator(
volume ** (1 / 3),
volume ** (1 / 3),
volume ** (1 / 3),
)
# Set random seed for reproducibility
generator.set_seed(12345)
# Set mean bead-to-bead distance
generator.set_mean_squared_bead_distance(
params.get("<b^2>").to(params.get("distance_units") ** 2).magnitude
)
# Add strands
generator.add_strands(
nr_of_strands=n_strands, # 10000 polymer chains
strand_lengths=[
n_atoms_per_strand for _ in range(n_strands)
], # 100 beads per chain
)
# At this point, the universe is still a melt
universe = generator.get_universe()
# Sample vulcanization "crosslinks"
sampled_crosslinks = randomly_sample_entanglements(
universe, nr_of_samples=int(0.75 * n_strands), upper_cutoff=2.5, seed="12345"
)
n_sampled_crosslinks = len(sampled_crosslinks.pairs_of_atoms)
# Add sampled crosslinks to the universe
universe.add_bonds(
n_sampled_crosslinks,
[sampled_crosslinks.pairs_of_atoms[i][0]
for i in range(n_sampled_crosslinks)],
[sampled_crosslinks.pairs_of_atoms[i][1]
for i in range(n_sampled_crosslinks)],
[2 for _ in range(n_sampled_crosslinks)], # Bond type 2
)
# Set the crosslinker type to 2 (vulcanization)
for i in range(n_sampled_crosslinks):
universe.set_vertex_property(
sampled_crosslinks.pairs_of_atoms[i][0], "type", 2)
universe.set_vertex_property(
sampled_crosslinks.pairs_of_atoms[i][1], "type", 2)
print("Generated vulcanized network with {} crosslinks".format(n_sampled_crosslinks))
# if you want, you can collapse the crosslinks to be a single atom
# as such:
universe = universe.contract_vertices_along_bond_type(2)
# Save the universe to a file
writer = DataFileWriter(universe)
if not os.path.exists("generated_structures"):
os.makedirs("generated_structures")
writer.write_to_file("generated_structures/vulcanized_network.data")
Generated vulcanized network with 7500 crosslinks
Refer to File I/O: Readers & Writers for more details on how you can save the universe to a file.
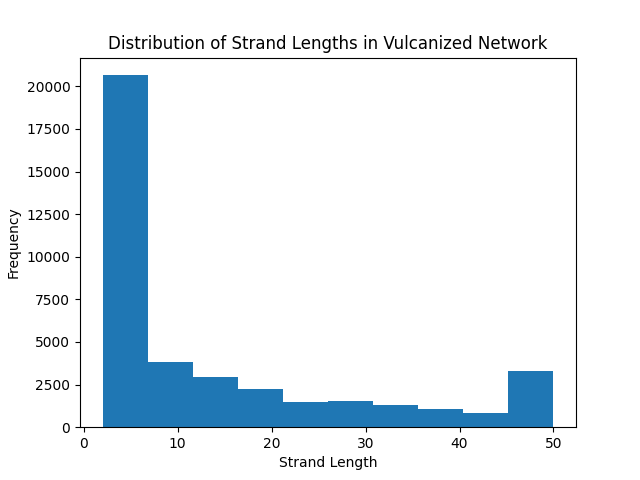
For now, we will show that this produces a variety of different strand lengths:
strand_lengths = [m.get_nr_of_atoms()
for m in universe.get_chains_with_crosslinker(2)]
# Plot the distribution of strand lengths
plt.figure()
plt.hist(strand_lengths, label="Strand Lengths")
plt.xlabel("Strand Length")
plt.ylabel("Frequency")
plt.title("Distribution of Strand Lengths in Vulcanized Network")
plt.show()
Total running time of the script: (1 minutes 25.424 seconds)