Note
Go to the end to download the full example code.
Extract Synthesis Parameters¶
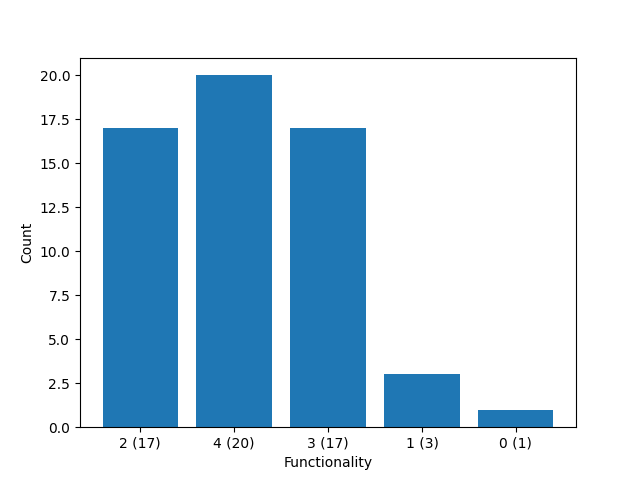
This example demonstrates how to analyze the structure of a crosslinked polymer network to get the stoichiometric imbalance \(r\), the crosslinker conversion \(p\) or the functionality of the crosslinker \(f\).
Stoichiometric imbalance (r): 1.16
Crosslinker conversion (p): 0.7241379310344828
Functionality of the crosslinker (f): 4
import os
from collections import Counter
import matplotlib.pyplot as plt
from pylimer_tools.analyse_networks import (
compute_crosslinker_conversion,
compute_stoichiometric_imbalance,
)
from pylimer_tools.io.read_lammps_output_file import read_data_file
from pylimer_tools_cpp import Universe
# Replace with your crosslinker type
crosslinker_type = 2
# Load your network (replace path accordingly)
universe = read_data_file(
os.path.join(
os.getcwd(),
"../..",
"tests/pylimer_tools/fixtures/structure/network_100_a_46.structure.out",
)
)
assert isinstance(universe, Universe)
# 1. Compute stoichiometric imbalance
r = compute_stoichiometric_imbalance(universe, crosslinker_type)
print(f"Stoichiometric imbalance (r): {r}")
# 2. Compute crosslinker conversion
p = compute_crosslinker_conversion(universe, crosslinker_type)
print(f"Crosslinker conversion (p): {p}")
# 3. Compute functionality of the crosslinker
f = universe.determine_functionality_per_type()[crosslinker_type]
print(f"Functionality of the crosslinker (f): {f}")
# Count functionalities
crosslinkers = universe.get_atoms_by_type(crosslinker_type)
functionalities = universe.get_vertex_degrees()
functionalities = [
functionalities[universe.get_vertex_idx_by_atom_id(a.get_id())]
for a in crosslinkers
]
functionality_counts = Counter(functionalities)
# Plot the functionality counts
plt.figure()
plt.bar(
[f"{k} ({v})" for k, v in functionality_counts.items()],
list(functionality_counts.values()),
)
plt.xlabel("Functionality")
plt.ylabel("Count")
plt.show()
Total running time of the script: (0 minutes 0.063 seconds)