Note
Go to the end to download the full example code.
Custom Crosslinking with Callback Functions¶
This example demonstrates how to use the link_strands_callback()
method to create custom crosslinking procedures
as an alternative to the built-in linking methods,
link_strands_to_conversion() and
link_strands_to_soluble_fraction().
The callback function allows users to control when the crosslinking process should stop based on
any criteria they can implement.
The callback function receives two parameters: - The current MCUniverseGenerator instance - The current step number
And should return one of the BackTrackStatus values: - STOP: Stop the linking process - TRACK_FORWARD: Continue linking forward - TRACK_BACKWARD: Track backward in the linking process
import math
import matplotlib.pyplot as plt
from pylimer_tools.calc.miller_macosko_theory import predict_gelation_point
from pylimer_tools.io.bead_spring_parameter_provider import (
ParameterType,
get_parameters_for_polymer,
)
from pylimer_tools_cpp import BackTrackStatus, MCUniverseGenerator, MEHPForceBalance2
Example 1: Stop at Conversion OR Soluble Fraction¶
Using a callback allows for more complex logic, such as stopping the process based on multiple criteria as illustrated here.
# Get parameters for PDMS polymer density and bead distance
params = get_parameters_for_polymer(
"PDMS", parameter_type=ParameterType.GAUSSIAN)
n_xlinks = 500 # Number of crosslinkers
n_strands = 1000 # Number of strands
strand_length = 50 # Length of each strand
n_atoms_total = n_strands * strand_length + n_xlinks
volume = n_atoms_total / params.get_bead_density()
generator = MCUniverseGenerator(
volume ** (1 / 3),
volume ** (1 / 3),
volume ** (1 / 3),
)
generator.set_seed(42)
# Add crosslinkers and strands
generator.add_crosslinkers(n_xlinks, 4, 2) # functionality 4
generator.add_strands(n_strands, [strand_length] * n_strands, 1)
target_conversion = 0.9 # Stop at 95% conversion
target_wsol = 0.2 # Stop if soluble fraction is below 20%
# record data throughout the process
conversions = []
wsol_values = []
steps = []
def conversion_callback(gen, steps_remaining):
"""
Callback function to monitor crosslinking progress.
This function checks the current crosslinker conversion and soluble fraction,
and decides whether to stop the linking process.
.. todo:
Implement the search in a binary search fashion to improve efficiency
"""
step = n_strands * 2 - steps_remaining
assert isinstance(gen, MCUniverseGenerator), (
"Callback must receive a MCUniverseGenerator instance"
)
current_conversion = gen.get_current_crosslinker_conversion()
conversions.append(current_conversion)
steps.append(step)
if current_conversion >= predict_gelation_point(r=1, f=4, b2=1):
prev_wsol = wsol_values[-1] if wsol_values else 1.0
# Check if we need to run force balance
if (step % 10 == 0) or (
math.isclose(prev_wsol, target_wsol, rel_tol=1e-1, abs_tol=5e-2)
):
fb2 = gen.get_force_balance2()
assert isinstance(fb2, MEHPForceBalance2), (
"Force balance must be MEHPForceBalance2"
)
fb2.run_force_relaxation()
current_wsol = fb2.get_soluble_weight_fraction()
else:
current_wsol = prev_wsol
else:
# skip running force balance while
# we don't even expect to have a network
current_wsol = 1.0
wsol_values.append(current_wsol)
if current_conversion >= target_conversion or current_wsol <= target_wsol:
print(
" Target conversion {:.1f} or soluble fraction {:.1f} reached at step {}.".format(
target_conversion, target_wsol, step
)
)
return BackTrackStatus.STOP
else:
return BackTrackStatus.TRACK_FORWARD
generator.link_strands_callback(conversion_callback, 1.0)
final_conversion = generator.get_current_crosslinker_conversion()
print(
f" Final conversion: {final_conversion:.3f} and w_sol: {wsol_values[-1]:.3f}")
Target conversion 0.9 or soluble fraction 0.2 reached at step 1473.
Final conversion: 0.736 and w_sol: 0.198
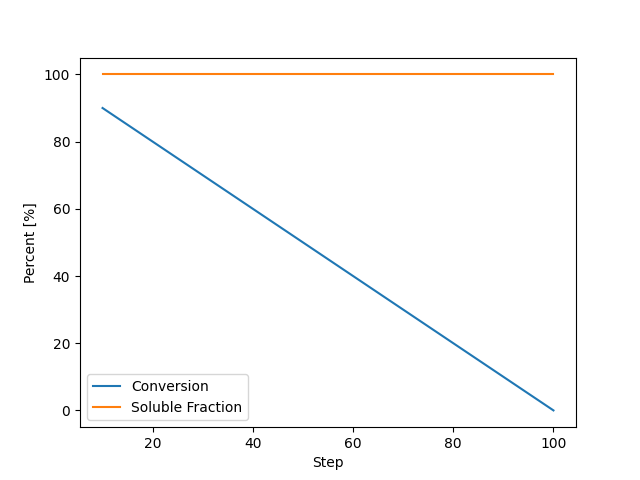
Visualizing the Results¶
Let’s plot the evolution of crosslinker conversion and soluble fraction over the steps.
fig, ax = plt.subplots()
ax.plot(steps, [c * 100 for c in conversions], label="Conversion")
ax.plot(steps, [wsol * 100 for wsol in wsol_values], label="Soluble Fraction")
ax.set_xlabel("Step")
ax.set_ylabel("Percent [%]")
ax.set(xlim=(0, max(steps)), ylim=(0, 100))
ax.legend()
plt.show()
Total running time of the script: (0 minutes 0.883 seconds)