Note
Go to the end to download the full example code.
Polydispersity Study¶
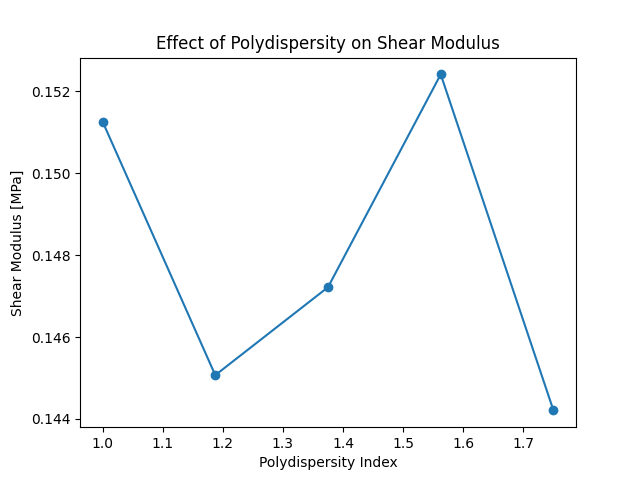
In this example, we study the effect of polydispersity on the shear modulus of end-linked polymer networks.
Again, please be aware that this is a minimal example, and the results are not statistically significant, because of the small system size and no repeated sampling of the polydispersity distribution nor entanglements.
import math
import random
import matplotlib.pyplot as plt
import numpy as np
from pylimer_tools.io.bead_spring_parameter_provider import (
ParameterType,
get_parameters_for_polymer,
)
from pylimer_tools_cpp import MCUniverseGenerator, MEHPForceBalance2
# Get parameters for PDMS polymer density and bead distance
params = get_parameters_for_polymer(
"PDMS", parameter_type=ParameterType.GAUSSIAN)
# setup strand lengths
n_strands = 10000
average_strand_length = 50
polydispersity = np.linspace(1.0, 1.75, 5) # Varying polydispersity
shear_moduli = []
for d in polydispersity:
s = math.sqrt(math.log(d))
m = math.log(average_strand_length) - 0.5 * s**2
# sample strand lengths from log-normal distribution
strand_lengths = [
int(random.lognormvariate(m, s)) if d > 1 else average_strand_length
for _ in range(n_strands)
]
n_atoms_total = (
sum(strand_lengths) + 0.5 * n_strands
) # also count crosslinker sites
volume = params.get_bead_density() * n_atoms_total
generator = MCUniverseGenerator(
volume ** (1 / 3),
volume ** (1 / 3),
volume ** (1 / 3),
)
generator.set_bead_distance(
params.get("<b>").to(params.get("distance_units")).magnitude
)
generator.add_strands(
nr_of_strands=n_strands,
strand_lengths=strand_lengths)
# 4-functional crosslinkers
generator.add_crosslinkers(int(0.5 * n_strands), 4)
generator.link_strands_to_conversion(
crosslinker_conversion=0.925, # 92.5% of crosslinker sites used
)
universe = generator.get_universe()
fb = MEHPForceBalance2(
universe,
nr_of_entanglements_to_sample=int(
params.get_entanglement_density() * universe.get_volume()
),
upper_sampling_cutoff=params.get_sampling_cutoff(),
entanglements_as_springs=False,
)
fb.run_force_relaxation()
# apply conversion factors, measure resulting shear modulus
r02_slope = params.get("R02")
r02_slope_magnitude = r02_slope.to(
params.get("distance_units") ** 2).magnitude
kbt = params.get("T") * params.get("kb")
gamma_conversion_factor = (
(kbt / ((params.get("distance_units")) ** 3)).to("MPa").magnitude
)
shear_modulus = (
gamma_conversion_factor
* np.sum(fb.get_gamma_factors(r02_slope_magnitude))
/ universe.get_volume()
)
shear_moduli.append(shear_modulus)
# Plot the results
plt.figure()
plt.plot(polydispersity, shear_moduli, marker="o")
plt.xlabel("Polydispersity Index")
plt.ylabel("Shear Modulus [MPa]")
plt.title("Effect of Polydispersity on Shear Modulus")
plt.show()
There are many things you would want to do differently in a real study, for example:
Use larger systems to get statistically significant results.
Sample the polydispersity distribution, the entanglements and the connectivity multiple times.
Change the polymer type and network properties as desired to match the experimental system.
Save the universe to a file for reproducibility and further analysis.
Total running time of the script: (5 minutes 41.163 seconds)